Nankai University Research Team Developes High-Affinity Neutralizing Antibody from SARS-CoV-2

Guo Yu, Zhang Hongkai, and Rao Zihe and their joint research team of Nankai University, successfully isolated neutralizing antibodies P4A1, P20A2 and P20A3 from patients recovered from Covid-19 in collaboration with Xinhua Hospital of Shanghai Jiao Tong University, Wuhan Institute of Virology of CAS, and HiFiBiO Therapeutics (Hangzhou). With the high-resolution structural information provided by X-ray crystallography, they explained the pharmacological mechanism of the neutralizing antibody P4A1. The antibody demonstrated high efficacy in treating a rhesus monkey carrying SARS-CoV-2.
On May 11, the paper A SARS-CoV-2 neutralizing antibody with exceptional spike binding coverage and optimized therapeutic potentials was published in Nature Communications, a peer-reviewed, open access, scientific journal.
The research team firstly used the spike protein of SARS-CoV-2 as a bait, isolating B lymphocytes (or B cells) from peripheral blood mononuclear cells in the recovered Covid-19 patients; then they examined the sequence information from B lymphocytes combined with the spike protein by applying single cell sequencing (NGS) technologies. As a result, three antibodies with high affinity, P4A1、P20A2 and P20A3, were acquired. All the three antibodies can efficiently bind the S1 subunit of the spiked protein while inhibiting the binding of the S1 subunit to the viral invasion receptor ACE2. Besides, in both neo-coronavirus and live virus neutralization assays, the antibodies all showed good neutralizing activity.
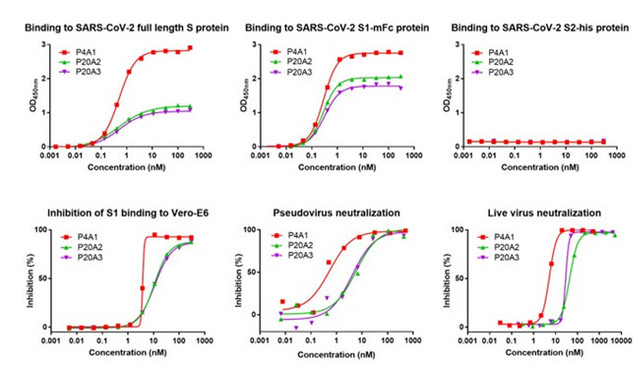
Figure 1 In vitro biological activity assay of three neutralizing antibodies
In order to investigate the molecular mechanism of the neutralizing antibody; the team obtained the antibody P4A1 at a resolution of 2.1 Å and its complex with the receptor binding domain (RBD) of the SARS-CoV-2 spike glycoprotein by X-ray crystallography. The results showed that three complementary-determining regions (CDRs) on the heavy chain of P4A1, two on the light region, and a light chain framework region 3 (LFR3) together form the interaction for binding to the spike protein. Their binding epitopes cover most of the recognition epitopes of the cellular receptor ACE2 on the spike protein, among which 15 out of 17 ACE2-recognized amino acids were occupied. Sequentially, it prevented the RBD of the SARS-CoV-2 spike protein from binding to the invasion receptor ACE2, and thus blocking the viral invasion.
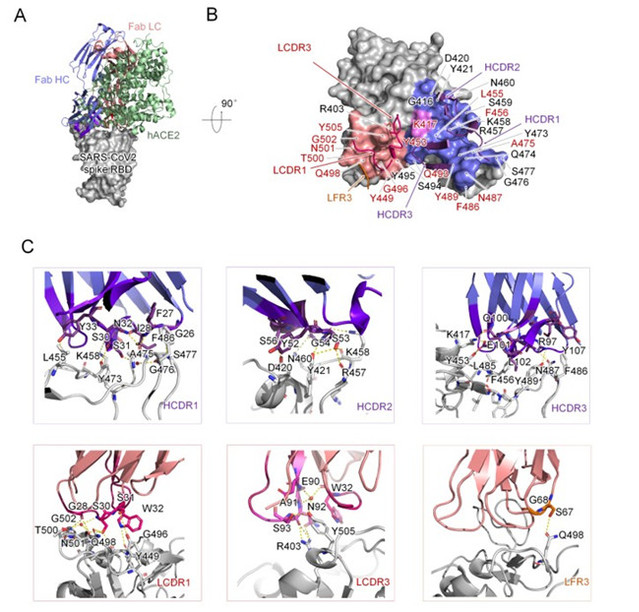
Figure 2 Complex Structure of P4A1 with SARS-CoV-2 spike protein receptor binding domain (RBD)
What is worth noting here is, the research teams applied antibody engineering and modification technologies to meet various clinical requirements and improve clinical utilities for antibody P4A1. That means to modify IgG1 to IgG4 to reduce the risk of antibody-dependent potentiation reported in multiple coronaviruses and significantly extend the plasma half-life of the antibody, providing long-term protection for individuals while increasing safety.
On this basis, the researchers teamed up with Shan Chao and Yuan Zhiming's team of Wuhan Institute of Virology of CAS to validate the therapeutic effect of antibodies on a rhesus monkey carrying SARS-CoV-2. The results revealed that the control group had severe bronchopneumonia with alveolar cavity edema and massive lymphocytic infiltration and bronchial epithelial cell detachment in the lungs. The antibody treatment group was able to effectively reduce the viral load in oropharyngeal and organ tissues, which greatly alleviates the symptoms of bronchopneumonia, and therefore protect the normal alveolar cavity.

Figure 3 Histopathological study of the lung in rhesus monkey model
Thesis link: https://www.nature.com/articles/s41467-021-22926-2
(Reported by Junhui Wu, translated by Meixu Wu, edited by Davide Francolino and JianjingYun)









