Nankai Research Team Makes Important Progress in Predicting Gene Expression and Interpreting Non-coding Variation
Recently, the team of Zeng Wanwen, a lecturer of College of Software, Nankai University, has made important progress in predicting gene expression and interpreting non-coding variation. The research results were published in Nature Machine Intelligence, a sub-journal of Nature with impact factor of 15.508,under the title "Reusability report: Compressing regulatory networks to vectors for interpreting gene expression and genetic variants" .
The genome-wide association study (GWAS) is an effective research strategy for identifying risk genes/loci of complex diseases, which points out the direction for the research on complex diseases, lays a solid foundation for the realization of personalized diagnosis, prognosis, and treatment, and promotes the development of human genetics and genomics research. However, most of the risk loci identified in GWAS are located in non-coding regions, and it is unclear whether these loci are directly related to disease functions. Accurate interpretation of the loci in the non-coding regions of GWAS is a vital prerequisite for achieving accurate medical treatment.
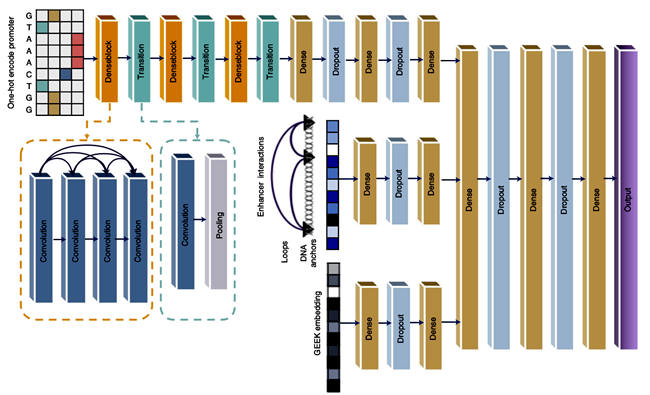
To systematically analyze the impact of the regulatory network on non-coding genetic variation, the research team further integrated the sequence data, HiChIP 3D genomic data, and low dimensional embedding of the regulatory network data of the GEEK model based on the Deep Expression model previously developed by the team to improve the prediction ability of gene expression. The downstream research results have shown that the integration of sequence data, 3D genomic data, and regulatory network data can better understand the transcriptional regulation mechanism and the function of non-coding variation, which provides a new approach for more accurate interpretation of genetic variation of GWAS.
The College of Software, Nankai University, is the first unit of this paper. Zeng Wanwen, a lecturer at the College of Software, and Xin Jingxue, a postdoctoral fellow of Stanford University, are the co-first authors. Jiang Rui, a long-tenured associate professor of Tsinghua University and Wang Yong, a researcher of the Academy of Mathematics and Systems Science, CAS are the co-corresponding authors of this paper. The research has been supported by the National Natural Science Foundation of China and the National Key Research and Development Young Scientist Program of China.
Link to the paper: Https://www.nature.com/articles/s42256-021-00371-6
(Edited and translated by Nankai News Team)









