Academician Xu Jianguo's Team Made Progress in Reverse Zoonosis Study
It is founded that humans can transmit Shigella flexneri to cattle
Recently, the study on human-to-bovine transmission of Shigella flexneri by the team led by Xu Jianguo, an Academician of the Chinese Academy of Engineering, was published online in the journal Emerging Microbes & Infections. The team works under the State Key Laboratory for Infectious Disease Prevention and Control of the Chinese Center for Disease Control and Prevention as well as the Research Institute of Public Health, Nankai University. It is the first time in the world that scientists have found humans can transmit Shigella to yaks, beef cattle, dairy cattle, etc.
For many years, the academic community has agreed that Shigella is a pathogenic bacterium unique to humans, which means that human is the only host for such bacterium. It is often said that more than 70% of new infectious diseases come from animals. And the findings of this study is enlightening for how emerging infectious diseases occur and how they cause diseases in animals.

The team of Academician Xu Jianguo cooperated with multiple teams including the Suzhou Institute of Systems Medicine of the Chinese Academy of Medical Sciences, Beijing Normal University, the University of New South Wales, Australia, and Hebei Engineering University and performed genome sequencing of 34 animal Shigella flexneri strains isolated from yaks, dairy cattle and beef cattle, and 268 Shigella flexneri strains isolated from patients in 17 provinces and cities in China from 1950 to 2017. Phylogenetic analyses were then carried out with the genome sequences of 346 Shigella flexneri strains in the global public database. The study found that the 648 strains of Shigella flexneri could be divided into 7 PG groups (as shown in the figure). The 34 strains of animal Shigella flexneri could be divided into two groups, PG1 and PG3, with each group accounting for 52.94% (18/34) and 47.06% (16/34) respectively of the total. From the phylogenetic information, the strains of PG1 group appeared earlier, mainly isolated from yaks while the PG3 strains appeared later, mainly isolated from beef cattle and dairy cattle. The strains isolated from Chinese patients were mainly within PG3 group, accounting for 91.54% (249/272). There are obvious differences between PG1 and PG3 strains in terms of virulence plasmids and antimicrobial resistance genes. The PG3 strains isolated from animals are similar to the popular strains in the Chinese population in recent years, both of which are ST91 type with multi-drug resistance characteristics. Some of PG3 strains are Serotype X Variants newly emerged in recent years.
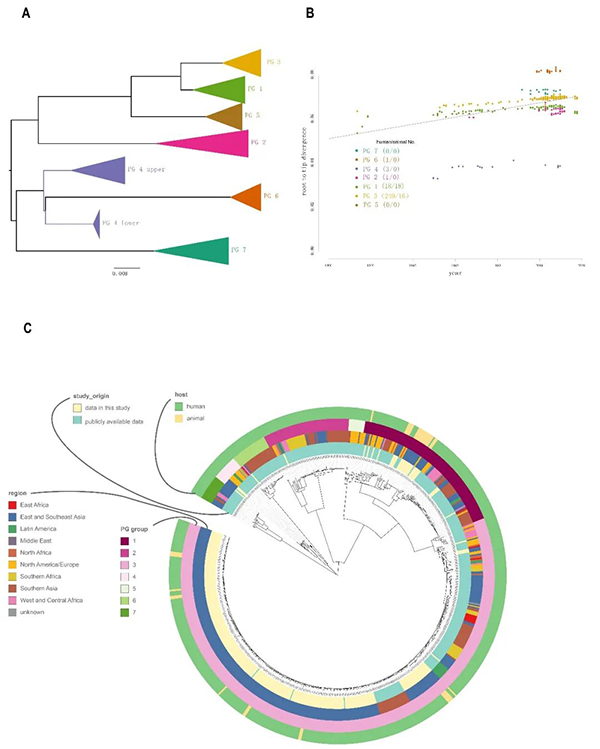
Figure: Population structure of 648 Shigella flexneri strains
PG1 population emerged as earliest as in around 1660 while PG3 population emerged as earliest as in about 1848. In the phylogenetic tree of the PG1 and PG3 strains, the strains isolated from humans are in the ancestral position, which fully suggests that Shigella was transmitted from humans to cattle. BEAST software was used to conduct phylogenetic analysis and differentiation time estimation of Shigella flexneri populations from different hosts. The results showed that PG1 animal strains can be divided into 4 lineages with the emergence time estimated to be between 1948 and 1966, whereas, PG3 strains can be divided into 8 lineages with the emergence time estimated to be between 1997 and 2016. That is to say, the above time periods were when Shigella flexneri was transmitted from humans to cattle, and such transmission happened many times. After Shigella flexneri was transmitted from humans to cattle, the evolution rate of Shigella flexneri populations was significantly slowed down.
The limitations of this study are that only small number of animal strains are isolated and the collection time lasts short, so that the results of the analysis may be biased. For pathogens that have adapted to animals, some behavioral barriers must be overcomed by them before becoming human pathogens, that is, increasing the chance of contact with humans. Fortunately, no cases of Shigella flexneri reinfecting humans have been found so far.
The significance of this study is that given that many infectious diseases are behavioral and ecological infectious diseases, human activities, especially large-scale economic and social activities, will lead to the transmission of pathogens from animals to humans, causing epidemics among humans, such as SARS; meanwhile, pathogens can be transmitted from humans to animals, resulting in infectious diseases among animals.
The paper titled Evolutionary and genomic insights into the long-term colonization of Shigella flexneri in animals was published in Emerging Microbes & Infections (TOP1 Sector, Impact Factor: 19.568) in August 2022 under the joint efforts of 4 teams in China and abroad. The co-first authors include Dr. Liang Junrong from the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention; Dr. Zhu Zhen from Hebei University of Engineering, Professor Lan Ruiting from University of New South Wales; Dr. Meng Jing from the Suzhou Institute of Systems Medicine, Chinese Academy of Medical Sciences, and Dr. Bram Vrancken from the Katholieke Universiteit Leuven in Belgium. The co-responsible authors are Academician Xu Jianguo and research fellow Jiang Taijiao.
Obtain full text from the link: https://doi.org/10.1080/22221751.2022.2109514
(Edited and translated by Nankai News Team)









