NKU-STU Joint Team Reveals Three-dimensional Structure of new Mycobacterium Tuberculosis Target to Aid the development of New Anti-tuberculosis Drugs
On March 15, the team of Prof. Rao Zihe of Nankai University and the team of Prof. Zhang Lu of ShanghaiTech University published online their joint research findings in Nature Microbiology, an authoritative journal in the field of microbiology. They reported the first three-dimensional cryogenic electron microscopy (cryo-EM) structures of the membrane-bound phosphoribosyltransferase Rv3806c, a new drug target of Mycobacterium tuberculosis, a deadly pathogen that causes global human Tuberculosis. This result reveals the fine 3D structure of complex resulting from Rv3806c’s binding with its receptor substrate and donor substrate, respectively, and reveals the molecular mechanism by which the protein catalyzes phosphoribosyl transfer on the bacterial plasma membrane, providing an important theoretical basis for the study of Rv3806c as a new target for the development of targeted drugs.
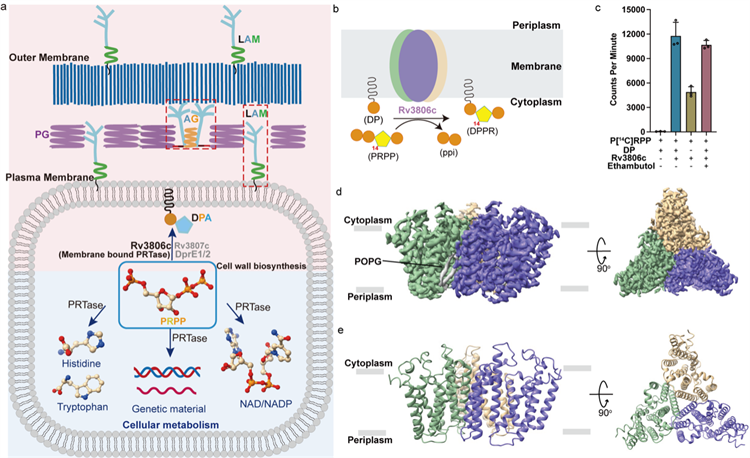
Figure 1 Functions and overall structure of Rv3806c
(a) Schematic diagram of the use of PRPP in Mtb in cell metabolism and cell wall biosynthesis.
(b) Schematic diagram of the reaction of Rv3806c to catalyze phosphoribosyl transfer on cell membrane.
(c) Radiolabeled PRTase in vitro enzyme activity experiment.
(d,e) Cryo-EM potential density plot and cartoon model of Rv3806c trimer.
The research team resolved the structure of Rv3806c’s two complexes with the acceptor substrate DP and the donor substrate PRPP. Cryo-EM results showed that Rv3806c was assembled in the form of trimer in a lipid environment. Subsequently, functional experiment revealed that the trimeric assembly form of Rv3806c plays an important role in performing the function of PRTase, and also confirmed the existence of Rv3806c in the form of trimer on the natural cytoplasmic membrane of mycobacteria. Each monomer is composed of 9 transmembrane helices (TM helices). TM 1-5 and TM 6-9 form two helical bundles respectively. The helices are connected by 4 periplasmic loops (PLs) and 4 cytoplasmic loops (CLs), while CL1-3 forms a cap structure above the transmembrane region. Structural and functional studies revealed that two helical bundles and a Cytoplasmic cap structure were responsible for binding substrates. The acceptor substrate DP binds to the hydrophobic cavity of the helical bundle-2, while the donor substrate PRPP binds to the cavity of the TM helical bundle-1 through one Mg2+. Binding affinity experiment showed that Mg2+ and ribose groups were essential for PRPP binding. In vitro PRTase enzyme activity assay showed that β-phosphate and terminal 5′-phosphate on PRPP played a key role in Rv3806c activity.
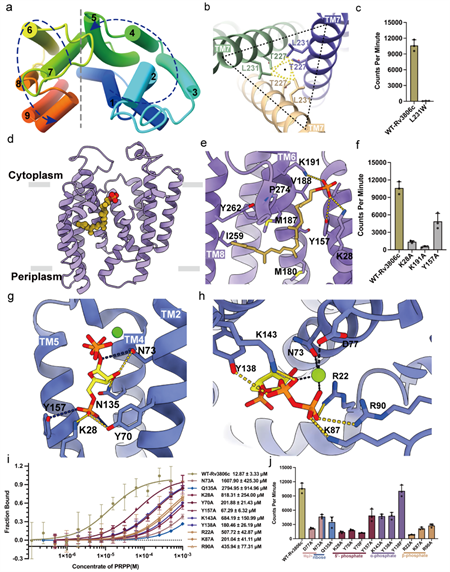
Figure 2. Detailed complex structure and function of Rv3806c
(a) Counterclockwise TM arrangement of Rv3806c, consisting of two helical bundles.
(b) TM7-mediated trimer interface and interacting amino acids.
(c) Results of PRTase in vitro enzyme activity experiment of amino acid mutants at the trimer interface.
(d-e) Schematic diagram of receptor substrate DP binding to the cavity of helical bundle-2 and interacting amino acids.
(f) PRTase in vitro enzyme activity results of amino acid mutants at DP binding site.
(g-h) Schematic diagram of receptor substrate PRPP binding to the cavity of the TM helical bundle-1 and the binding site.
(i-j) Binding affinity of amino acid mutants at PRPP binding sites and results of PRTase in vitro enzyme activity.
Through the comparison of the two structures, it was found that the binding of PRPP led to different degrees of conformational changes in CLs, and this further closed the cap structure domain located on the cytoplasmic side, thereby forming the active center of the reaction and preventing the flow of solvents in the cytoplasm to the catalytic center. This is also a general strategy adopted for this family of enzymes, including soluble PRTases. By superimposing and comparing the structures of the two complexes, it was found that the oxygen atom of phosphoric acid in DP was close to the ribose anomeric carbon C1 atom of PRPP (4.5 Å). The phosphoric acid in DP is located opposite the leaving group PPi, and the phosphoric acid that satisfies the nucleophile DP attacks the ribose C1 atom of PRPP from the back, that is, “inverting” nucleophilic substitution. This reaction converts the α configuration of the donor PRPP ribose into the β configuration of the product DPPR ribose.
This work also explained the mechanism of clinical mutation on Rv3806c that causes ethambutol resistance through structural and functional studies, suggesting that the phospholipid binding site located on the trimeric interface affects the PRTase of the trimer through a possible allosteric regulation mechanism, thereby mediating clinical ethambutol resistance.
According to introduction, Gao Shan, a 2022 cohort doctoral student from the College of Life Sciences of Nankai University, and Wu Fangyu, a 2020 cohort doctoral student from the College of Life Sciences, are the first authors of the paper. Nankai University is the primary affiliation of the paper. Prof. Zhang Lu from the Shanghai Institute for Advanced Immunochemical Studies, academician Rao Zihe and Fellow Gurdyal S. Besra from the University of Birmingham are the co-corresponding authors of the paper.
Electron microscopy data collection was completed in the Bio-EM Center of ShanghaiTech University. This work is another important achievement in the synthesis of new targets for anti-tuberculosis cell wall by the joint research team of Zhang Lu / Rao Zihe and Gurdyal S. Besra from the University of Birmingham after revealing the molecular mechanism of the target protein EmbA/B/C of ethambutol, a first-line anti-tuberculosis drug (Science, 2020) and explaining the fine 3D structure of AftA, a key enzyme in the AG synthesis pathway that initiates arabinose synthesis (PNAS, 2023). This provides a structural basis for a systematic understanding of the molecular mechanism of Mtb’s unique and complex cell wall assembly as well as the discovery of new anti-tuberculosis drugs targeting the cell wall.
Link: https://www.nature.com/articles/s41564-024-01643-8
Links to related achievements:
https://www.science.org/doi/10.1126/science.aba9102
https://www.pnas.org/doi/10.1073/pnas.2302858120
(Edited and translated by Nankai News Team.)









