Nankai University Research Team Reports Significant Breakthrough in RNA m⁶A Modification
On November 29, the research group led by Changliang Shan at the College of Pharmacy, Nankai University (NKU), published a paper in Advanced Science with the title HPD is an m⁶A Methyltransferase that Protects Colorectal Cancer Cells from Ferroptotic Cell Death by m⁶A Methylating SLC7A11/GPX4. In this study, Shan Group discover the tyrosine metabolism enzyme 4-hydroxyphenylpyruvate dioxygenase (HPD) as a methyltransferase responsible for m6A modification. Unlike METTL3, which requires the assistance of METTL14 to form a complex and exert methyltransferase activity. Interestingly, we reveal that HPD has a catalytic domain (CMI) like METTL3. Moreover, HPD recruits the universal cofactor S-adenosylmethionine (SAM) to the substrate binding center as a methyl group donor. In particular, we demonstrate that HPD regulates colorectal cancer ferroptosis by methylating SLC7A11/GPX4 through a moonlighting function. These findings uncover the moonlighting function of HPD in m6A-mediated ferroptosis and underscore the potential to target the m6A methyltransferase activity of HPD for cancer treatment.
4-hydroxyphenylpyruvate dioxygenase ( HPD ) is the second key enzyme in the tyrosine catabolism pathway, which catalyzes the conversion of 4-hydroxyphenylpyruvic acid (4-HPPA) to homogentisic acid (HGA).An increasing number of metabolic enzymes have been shown to have both non-enzymatic and multi-enzymatic activities. However, whether HPD has multi-enzymatic activities in regulating cancer development has not been reported. They discovered the presence of HPD in the nucleus. This finding suggests that HPD may possess previously unrecognized biological functions within the nucleus. Transcriptomic sequencing analysis of HPD-knockdown cells revealed significant enrichment of pathways related to RNA modification and RNA methylation. This finding provided key clues supporting this hypothesis. Coupled with our previous finding that HPD possesses RNA - binding capability, we propose that HPD is likely involved in RNA methylation processes within the nucleus.
To validate whether HPD regulates m⁶A methylation, we employed a combination of techniques, including dot blot, ELISA, and LC/MS, in both HPD-overexpressing and HPD-knockdown cells, as well as in HPD-knockout mouse models. These experiments confirmed that HPD positively regulates m⁶A modification. Given that METTL3 is a well-known m⁶A methyltransferase, we further investigated whether HPD's function is dependent on METTL3. We found that overexpression of HPD in METTL3-knockdown or knockout cells still resulted in an increase in m⁶A levels. More importantly, in an in vitro methylation assay, HPD alone was sufficient to catalyze m⁶A methylation on total RNA or single-stranded RNA. Collectively, these results demonstrate that HPD's catalysis and regulation of m⁶A modification are completely independent of METTL3. Moreover, HPD can function autonomously as a methyltransferase and exhibits more flexible catalytic properties compared to METTL3. Additionally, we identified key residues essential for this function, including the SAM - binding sites (H183 and H266) and the catalytic motif (CMI). We found that mutations at these sites significantly impaired HPD's methyltransferase activity.
This study also revealed that both a tyrosine metabolic inhibitor of HPD (Nitisinone) and its physiological substrate (4-hydroxyphenylpyruvate) could inhibit HPD's methyltransferase activity, both in cellular systems and in in vitro methylation assays. More importantly, mutations at the key sites we identified, which regulate its methyltransferase activity, also concurrently affected its intrinsic tyrosine metabolic enzyme activity. Through structural analysis and comparative assessment of the results, we confirmed that these two distinct catalytic functions of HPD share the same active site pocket. This discovery provides a crucial structural basis for the observed functional antagonism. Moreover, the C - terminal region of HPD is responsible for its RNA - binding activity. In summary, HPD is a multifunctional protein integrating metabolism, gene expression regulation, and RNA binding. In future studies aiming to precisely dissect its specific biological functions, it will be essential to carefully define or rule out the potential contributions of its other activities.
The ferroptosis pathway and its related inhibitors have emerged as a highly promising new direction in cancer therapy. Our sequencing analysis indicated that the ferroptosis pathway plays a pivotal role in the mechanism of action of HPD. Specifically, HPD influences cellular ferroptosis by regulating the expression of its core genes, SLC7A11 and GPX4. Furthermore, in - depth mechanistic exploration revealed that HPD enhances m⁶A methylation on SLC7A11 and GPX4 mRNA, thereby increasing their stability and ultimately promoting their protein expression. A series of experiments conducted on cellular, organoid, and animal models consistently demonstrated that targeted inhibition of HPD not only effectively reduces global m⁶A levels and SLC7A11/GPX4 expression but also significantly suppresses tumor growth. These findings highlight the substantial therapeutic potential of HPD as a novel target for colorectal cancer.
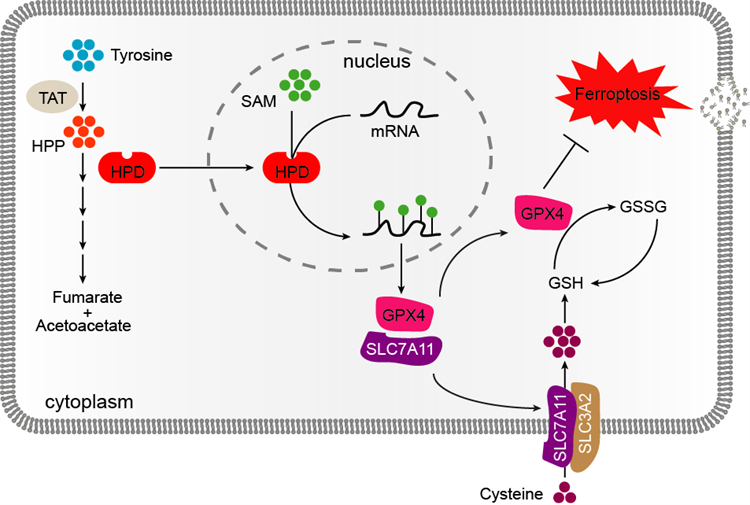
Schematic Illustration of HPD-Mediated Methylation of SLC7A11/GPX4 in Regulating CRC Ferroptosis and Tumor Growth
The co-first authors of the paper are Jiyan Wang (postdoctoral fellow), Xintong Dai (Ph.D. candidate, class of 2022), and Huanle Liu (Ph.D. candidate, class of 2022) from the NKU College of Pharmacy. The corresponding authors are Prof. Changliang Shan from the NKU College of Pharmacy, Researcher Shuai Zhang from the College of Integrative Chinese and Western Medicine, Tianjin University of Traditional Chinese Medicine, and Chief Physician Chunze Zhang from Tianjin People’s Hospital.
Read the paper at
https://advanced.onlinelibrary.wiley.com/doi/10.1002/advs.202508541
(Edited and translated by Nankai News Team.)









